News
Blasting Antibodies with Lasers Provides Direct Way of Measuring their Flexibilities
Published December 15, 2002
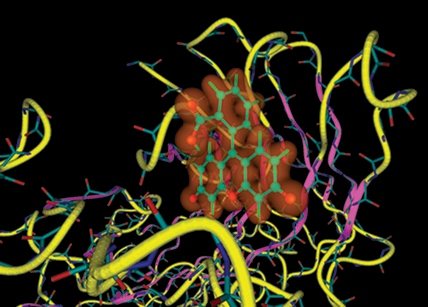
In this visualization of a fluorescein antigen bound to an antibody molecule, the primary protein structure is blue and red, secondary structures such as ribbons are purple and yellow. Fluorescein is green and its electrostatic cloud is orange. The visualization was created by a software program called QMView, which is integrated with a quantum chemistry software package called General Atomic and Molecular Electronic Structure System (GAMESS). |
A group of scientists at The Scripps Research Institute (TSRI) and San Diego Supercomputer Center (SDSC) at UCSD have used a powerful laser in combination with innovative quantum mechanical computations to measure the flexibility of mouse antibodies.
The new technique, described in an upcoming issue of the journal Proceedings of the National Academy of Sciences, is significant because protein flexibility is believed to play an important role in antibody-antigen recognition, one of the fundamental events in the human immune system.
"This is the first time anybody has ever gone into a protein and experimentally measured the frequency of vibrations in response to an applied force," said Floyd Romesberg, assistant professor in the Department of Chemistry at TSRI, who led the study.

These are two models of antibody-antigen binding: lock and key (above), and induced-fit (below).
|
Flexibility of Proteins
There are lots of ideas about mechanisms of antigen recognition postulated in the literature, but what the debate comes down to is really a question of flexibility. How flexible are proteins?
Unfortunately, flexibility is difficult to characterize experimentally, and there has never been a study like this one to carefully examine the details of antibody recognition of antigen at the molecular level, which involves bond vibrations that ever-so-slightly displace atoms a million times every millionth of a second. Scientists have had a tough time studying these vibrations because the two main techniques that allow them to "look" at proteins-nuclear magnetic resonance (NMR) and X-ray crystallography-cannot be used.
X-ray crystallography provides only average structures, which provide no direct information on a protein's flexibility. NMR can, in principle, be used to measure a protein's flexibility, but in practice is limited to slow timescales, involving large amplitude motions. Moreover, the number of atoms within a molecule the size of an antibody is so large that drawing conclusions from the data is nearly impossible.
Now Romesberg, Baldridge, and their colleagues have developed a way to measure and calculate the flexibility of proteins over timescales ranging from femtoseconds to nanoseconds using a combination of spectroscopy and quantum mechanical techniques.
Shaken, Not Stirred
For years biochemists have routinely used bench top ultraviolet and visible light spectrometers to measure things like protein concentration or to follow chemical reactions. However, Romesberg's spectrometer is not the kind you might find in any catalog of equipment lying around the lab.
This laser, built by Romesberg and Research Associate Ralph Jimenez, takes up an entire room and emits a burst of photons in a roughly 50-femtosecond pulse-which is billions of times faster than the fastest shutter speed on a good camera. This incredible speed is necessary, because just as a fast shutter speed captures a rapid movement on film, a fast laser captures a rapid movement within a protein.
"The laser allows us to take 'photographs' of a protein vibrating," said Romesberg.
In their experiments, Romesberg and Jimenez mix human antibodies with dye molecules. When the mixture is blasted with the laser beam, the dye molecules absorb energy from the laser and transmit some of this energy into the antibody.
The only place for the energy to go within the antibody is for it to be absorbed by vibrating bonds within the protein. The electron distribution in these bonds may then change, depending on how much they vibrate. By comparing an excited "spectra" readout to a normal spectrum, Romesberg and his colleagues can assess how flexible particular parts of a protein are.
This is not always simple. Antibodies are large proteins with lots of vibrating bonds, and molecular motions. Quantum mechanical calculations can help researchers delineate which motions are primary participants in the antibody-antigen recognition process. Baldridge, who performed such computations, then took the results and provided a visual way to understand the effect of the force on the protein.
The quantum mechanical results actually give a depiction of the electrostatic processes that are occurring. Together with the experimental information, this helps complete the puzzle of how various bonds are moving, twisting, and interacting with other atoms in the protein environment.
The lock-and-key model specifies that if the antigen and antibody are not matched up in a rigid, structural way, they will not bind. Romesberg, Baldridge, and their colleagues found this to be true for one three antibodies they tested. But the two other antibodies appeared to wiggle a lot to achieve their optimal energetic state.
Antibody recognition, said Romesberg, may not be a simple, lock-and-key mechanism, but one in which the keys and the locks are vibrating and changing their shape as they come together in solution.
The article, "Flexibility and Molecular Recognition in the Immune System" was authored by Ralph Jimenez, Georgina Salazar, Kim K. Baldridge, and Floyd E. Romesberg, and appears in the online edition of Proceedings of the National Academy of Sciences the week of December 16, 2002. The article will appear in print early next year.
This work was supported by the Skaggs Institute for Chemical Biology, the National Science Foundation, and the National Institutes of Health.
Media contacts:
Jason Bardi, (858) 784-9254, jasonb@scripps.edu
Rex Graham, (858) 822-5408, rgraham@sdsc.edu



