News
Mapping Tool Spots Brain Disorders
Published October 05, 2006
|
|
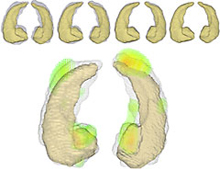
Using TeraGrid resources at SDSC and NCSA via GPFS-WAN, an advanced mapping tool developed by BIRN researchers was used to distinguish normal from diseased brain scans. The large image shows velocity vector information which highlights significant Hippocampal deformation areas or brain regions that differ between the control and Alzheimer's scans.
Credit: Timothy Brown, BIRN, CIS at Johns Hopkins University. Created with ParaView and MayaVi. |
by Paul Tooby,
SDSC Senior Science
Brain diseases are challenging to diagnose, which can confuse or delay beneficial treatments. Now, researchers in the Biomedical Informatics Research Network (BIRN) are using specific structural or shape differences in patients' brains to help identify brain disorders. "Using TeraGrid resources at multiple sites, this research has been able to successfully distinguish diagnostic categories such as Alzheimer's and Semantic Dementia from control subjects," said Anthony Kolasny of the Center for Imaging Science (CIS) at Johns Hopkins University . "This can potentially lead to a powerful new cyberinfrastructure tool clinicians can use to make earlier, more accurate diagnoses."
The BIRN Network, under the direction of UC San Diego Professor Mark Ellisman and funded by the National Institutes of Health/National Center for Research Resources, is an innovative and award-winning geographically distributed virtual community of shared resources offering tremendous potential to advance the diagnosis and treatment of disease.
The BIRN researchers at CIS and other participants in the Morphometry BIRN test bed have collaborated on a multi-institution computational anatomy processing pipeline that can handle the demanding analysis of brain structure data. The CIS shape-based morphometric tool, the Large Deformation Diffeomorphic Metric Mapping (LDDMM) tool, was used to study hippocampal data from 101 of subjects in three categories, Alzheimer's, Semantic Dementia, and control subjects. The collaborators include Michael Miller of Johns Hopkins University, Bruce Fischl of Massachusetts General Hospital, Randy Buckner of Harvard, and Carey Priebe and Anthony Kolasny of Johns Hopkins.
High resolution structural MRI brain scans at one BIRN site were segmented at a second BIRN site, then the data sets were accessed, aligned, and processed at the CIS site with the LDDMM tool using TeraGrid computing resources. The mapping tool computes a mathematical description of which shapes are similar and different by computing metric distances in the space of anatomical images, allowing direct comparison and quantitative characterization of differences in brain structure shapes.
Because the computations are demanding, requiring a large number of data-oriented jobs, the researchers wanted to run on resources beyond those of just one center. In a textbook case of TeraGrid use, and helped by an Advanced Support for TeraGrid Applications (ASTA) collaboration at the San Diego Supercomputer Center (SDSC), the researchers were able to access sufficient computing resources by running some 130,000 CPU hours on TeraGrid resources at both SDSC and the National Center for Supercomputing Applications (NCSA) in Illinois. This was feasible because the researchers were able to use the General Parallel File System-Wide Area Network (GPFS-WAN), pioneered by SDSC, to move 29 terabytes of output in some 4 million files between NCSA and their data store at SDSC.
"Users have always needed performance, and with complex computational infrastructures now in place, straightforward operation and reliability for data movement is essential," said Phil Andrews, SDSC's Program Director for High-End Computing Technologies. "GPFS-WAN makes grid computing at multiple sites accessible and efficient by providing users transparent, high-speed access to files across the grid."
Using GPFS-WAN allowed the researchers to maintain the principal copy of their data on SDSC's state-of-the-art data storage resources, for more reliable curation in accordance with modern data management practices. Then, via the Global File System they could directly access this data from multiple locations, efficiently running compute jobs at NCSA as well as SDSC. Before GPFS-WAN, to compute at a remote center the data had to be moved for every compute job, allowing the BIRN researchers to complete only about 70 jobs per day. But with GPFS-WAN the data is available on the Global File System and accessed directly from each site, increasing their throughput dramatically and helping them complete more than 1,000 jobs per day.
In addition to being able to compute efficiently across multiple TeraGrid sites, the researchers also mounted GPFS-WAN at their home institution of Johns Hopkins. Using sshfs (a filesystem client based on the SSH File Transfer Protocol) they ran ParaView visualization software locally at Johns Hopkins on their remote data at SDSC, giving them an effective method to explore the very large data sets produced by high-resolution brain scans, without incurring huge data transfer costs.

