News
Scientists Uncover Strong Support for Once-Marginalized Theory on Parkinson’s Disease
Published April 24, 2012
|
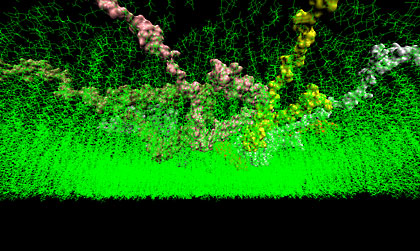
This image shows a construction of a possible ring oligomer position in the cell membrane after four nanoseconds of molecular dynamics simulations. Image courtesy of Igor Tsigelny, San Diego Supercomputer Center and Department of Neurosciences, UC San Diego. |
University of California, San Diego scientists have used powerful computational tools and laboratory tests to discover new support for a once-marginalized theory about the underlying cause of Parkinson’s disease.
The new results conflict with an older theory that insoluble intracellular fibrils called amyloids cause Parkinson’s disease and other neurodegenerative diseases. Instead, the new findings provide a step-by-step explanation of how a “protein-run-amok” aggregates within the membranes of neurons and punctures holes in them to cause the symptoms of Parkinson’s disease.
The discovery, published in the March 2012 issue of the FEBS Journal, describes how α-synuclein (a-syn), can turn against us, particularly as we age. Modeling results explain how α-syn monomers penetrate cell membranes, become coiled and aggregate in a matter of nanoseconds into dangerous ring structures that spell trouble for neurons.
“The main point is that we think we can create drugs to give us an anti-Parkinson’s effect by slowing the formation and growth of these ring structures,” said Igor Tsigelny, lead author of the study and a research scientist at the San Diego Supercomputer Center and Department of Neurosciences, both at UC San Diego.
Familial Parkinson’s disease is caused in many cases by a limited number of protein mutations. One of the most toxic is A53T. Tsigelny’s team showed that the mutant form of α-syn not only penetrates neuronal membranes faster than normal α-syn, but the mutant protein also accelerates ring formation.
“The most dangerous assault on the neurons of Parkinson’s patients appears to be the relatively small α-syn ring structures themselves,” said Tsigelny. “It was once heretical to suggest that these ring structures, rather than long fibrils found in neurons of people having Parkinson’s disease, were responsible for the symptoms of the disease; however, the ring theory is becoming more and more accepted for this neurodegenerative disease and others such as Alzheimer’s disease. Our results support this shift in thinking.”
The modeling results also are consistent with the electron microscopy images of neurons in Parkinson’s disease patients; the damaged neurons are riddled with ring structures.
Wasting no time, the modeling discoveries have spawned an intense hunt at UC San Diego for drug candidates that block ring formation in neuron membranes. The sophisticated modeling required involves a complex realm of science at the intersection of chemistry, physics, and statistical probabilities. A kaleidoscope of interacting forces in this realm makes α-syn proteins bump and tremble like they’re in an earthquake, coil and uncoil, and join together in pairs or larger groups of inventive ballroom dancers.
The modeling is creating a much better understanding of the mysterious a-syn protein itself, according to Tsigelny. A few years ago it was shown to accumulate in the central nervous system of patients with Parkinson’s disease and a related disorder called dementia with Lewy bodies.
The new modeling study has revealed precisely how two α-syn proteins insert their molecular toes into the membrane of a neuron, wiggle into it in only a few nanoseconds and immediately join together as a pair. The pair isn’t itself toxic; however, when more α-syn proteins join the dance, a key threshold is eventually crossed; polymerization accelerates into a ring structure that perforates the membrane, damaging the cell.
Tsigelny said many ring structures may be required to actually kill neurons, which are known for their durability. The nerve cells may be able to repair dozens of ring-induced perforations, keeping pace with a-syn assault. But at some point, the rate of perforations surpasses the ability of neurons to repair them. As a result, symptoms of Parkinson’s disease gradually appear and worsen.
“We think we can create a drug that stops the α-syn polymerization at the point of non-propagating dimers,” Tsigelny said. “By interrupting the polymerization at this crucial step, we may be able to slow the disease significantly.”
Tsigelny’s research team included Yuriy Sharikov, with SDSC and UC San Diego’s Department of Neurosciences; Wolfgang Wrasidlo, with the university’s Moores Cancer Center; and Tania Gonzalez, Paula A. Desplats, Leslie Crews, and Brian Spencer, all with UC San Diego’s Department of Neurosciences. The experimental validation studies were performed by Eliezer Masliah, a professor in the UC San Diego departments of Neurosciences and Pathology, and his associates. They relied on 3-D models of proteins, plus molecular dynamics simulations of the proteins, other modeling techniques and cell-culture experiments.
Given their deeper understanding of α-syn polymerization in neurons, they are now focused on understanding how monomers of α-syn stick to one another. Their search for drug candidates will include molecules that induce different conformations of α-syn proteins that are less inclined to stick together. Tsigelny said this effect, even if small, could reduce symptoms.
This computationally intensive approach includes an examination of the many possible three-dimensional arrangements of α-syn dimers, trimmers and tetramers. Pharmaceutical companies have used versions of the approach to develop drug candidates designed to bind to ‘anchor residues’ or ‘hot spots’ within target proteins. Algorithms assess in virtual experiments the theoretical ability of thousands of candidate drugs to bind to human proteins in the ever-expanding database of known 3-D structures of those proteins.
However, attempts to find drugs this way have generated promising candidates that fail in clinical trials with expensive regularity.
“Out of these failures we’ve come to appreciate that proteins change their shapes so often that what would appear to be a primary drug target may be present one nanosecond, gone the next, or it wasn’t relevant in the first place,” said Tsigelny, a physicist-turned-drug-designer.
Tsigelny’s approach takes advantage of classical drug-discovery algorithms, but adds additional analytical techniques to expand the search to include how a target protein’s conformations change in response to the forces operating on the scale of molecules.
“Sometimes, the drug-discovery models, despite being ‘nice looking,’ can be completely wrong,” Tsigelny said. “Scientists involved in drug discovery need to know when and to what extent to trust them. Even a slight shift in a cell’s environment can profoundly change the interactions of proteins with neighboring molecules. We think it’s realistically possible to design a drug to treat neurodegenerative diseases such as Parkinson’s disease and other diseases like diabetes with a more fundamental understanding of the proteins involved in those diseases.”
The research was funded by grants from the National Institutes of Health and Department of Energy, with computational support from Argonne National Laboratory’s IBM Blue Gene supercomputer as well as computational resources at SDSC.
About SDSC
As an Organized Research Unit of UC San Diego, SDSC is considered a leader in data-intensive computing and all aspects of ‘big data’, which includes data integration, performance modeling, data mining, software development, workflow automation, and more. SDSC supports hundreds of multidisciplinary programs spanning a wide variety of domains, from earth sciences and biology to astrophysics, bioinformatics, and health IT. With its two newest supercomputer systems, Trestles and Gordon, SDSC is a partner in XSEDE (Extreme Science and Engineering Discovery Environment), the most advanced collection of integrated digital resources and services in the world.
Media Contacts:
Rex Graham, SDSC Communications
858 232-2706 or ragraham@ucsd.edu
Jan Zverina, SDSC Communications
858 534-5111 or jzverina@sdsc.edu
Warren R. Froelich, SDSC Communications
858 822-3622 or froelich@sdsc.edu
Categories
Archive
Related Links
San Diego Supercomputer Center: http://www.sdsc.edu/
UC San Diego: http://www.ucsd.edu/
UC San Diego Department of Neurosciences: http://neurosciences.ucsd.edu/
UC San Diego Department of Pathology: http://pathology.ucsd.edu/
Moores Cancer Center UC San Diego: http://cancer.ucsd.edu/