News
New Understanding of ‘Copper Pump’ in Cells Could Prime Discovery of Anti-Cancer Drugs
Published May 11, 2012
A team of University of California, San Diego researchers has made new discoveries about a copper-transporting protein in the membranes of human cells that drug-discovery scientists can co-opt for the development of new anti-cancer drugs.
The findings, published May 9 as an online-first paper in Cell Biochemistry and Biophysics, describe how the copper transporter works as a biochemical pump to seize copper atoms outside of a cell and whisks the atoms through the otherwise impervious cell membrane into the cell cytoplasm. The same pump transports the platinum-containing drug cisplatin into cancer cells to help kill them. Igor Tsigelny, a research scientist at the university’s San Diego Supercomputer Center and Department of Neurosciences, is lead author of the paper.
|
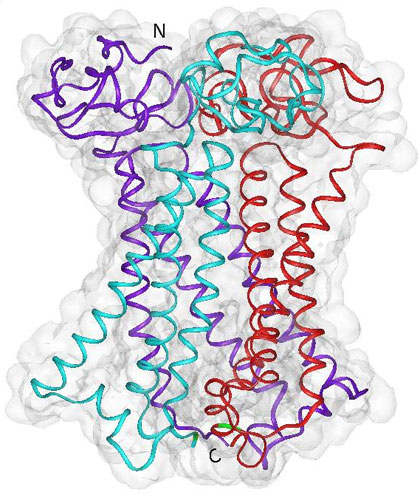
Researchers at UC San Diego used experimental results and modeling studies to discover that the human copper transporter protein forms a trimer (purple, aqua, and red) in a cell’s membrane, with one end (top) extending outside the cell and the other end (bottom) extending into the cell’s cytoplasm. Image courtesy of Igor Tsigelny, San Diego Supercomputer Center and Department of Neurosciences, UC San Diego. |
The body needs only a tiny amount of copper, but the little that is needed acts as a key component of vital cellular enzymes, including superoxide dismutase, cytochrome c oxidase, lysyl oxidase and dopamine β-hydrolase.
Researchers have shown before that that human copper transporter 1 (hCTR1) protein also participates in transport of the platinum-containing cisplatin, one of the most widely used anti-cancer drugs. Once platinum-containing cisplatin molecules enter a tumor cell, the molecules interact with the cell’s DNA and kill it in a process that has been extensively studied by Stephen B. Howell, a professor of medicine at the UC San Diego Moores Cancer Center.
The way that hCTR1 works is a focus of research by Howell and other cancer researchers because cisplatin and similar drugs somehow lose their punch: they are effective anti-cancer drugs when first administered, but lose much of their effectiveness during cancer relapses. Some researchers theorize that the diminished effect of cisplatin could be due to a change in hCTR1 in cancer cells.
New insights derived by the UC San Diego team is leading to a better understanding of what happens to the protein transporter and that knowledge could possibly be used to design a better version of cisplatin or an entirely new drug to take advantage of the new information.
In addition to cancer researchers, the hCTR1 has been a mystery to cell biologists. Until recently, they didn’t know whether the transporter protein formed dimers, or trimers. In a 2006 breakthrough that was refined in 2009, scientists confirmed that the trimer is the predominant structure, which was confirmed by the pioneering work of Northwestern University Professor Vincenz Unger.
Unger’s team identified the structure of the part of the hCTR1 transporter protein that spans the cell membrane. But they were not able to determine the structure of the part of the protein that extends to the outside of the membrane. Because of that gap in knowledge, they were not able to obtain a high-resolution 3-D map of the protein’s structure.
SDSC’s Tsigelny and his colleagues set out to create a complete, detailed 3-D model of the transporter. “There is no magic bullet in protein modeling, especially when we do not have a direct homologous template of another protein crystal structure,” Tsigelny said. “We predicted the structure of the protein on the level of information available at the current time, but this model needed to be checked with actual experimental results.”
Any model that Tsigelny’s team came up with would have to answer questions that had evaded scientists for years. For example, why is the extracellular end of the transporter so flexible? While the flexibility frustrated Unger’s ability to determine its 3-D structure, was the flexible tip of the protein stable enough to support its copper-transporting function?
Would the positively charged metal ions be transported electrostatically? And how does the transporter initially corral metal ions at pick-up points on the cell exterior and drop them off inside?
Tsigelny’s team used a computationally rigorous approach to find the answers.
So-called molecular dynamics modeling studies showed that the path the metal ions take through the intra-membrane transporter channel is stable despite the innate flexibility of the protein. In addition, while electrostatic forces worked well to hold positively charged metal ions like magnets at the extracellular and intracellular ends of the transporter protein, the passage of the metal atoms through an interior channel in the protein must be caused by another means.
Searching the Protein Data Bank
To help to understand the metals’ interaction with protein, Tsigelny’s team invented a new programming tool called METBIND, which works like a chemistry search engine. The program tried to find the possible binding sites of copper and platinum (along with other metal ions) as they interact with the hCTR1 protein and then move along it.
They checked the validity of their METBIND program with all possible copper-protein binding arrangements reported in the 74,000 proteins in the Protein Data Bank.
To the Tsigelny team’s surprise, the METBIND program correctly predicted 80 percent of all known copper binding sites in all 636 copper-binding proteins in the Protein Data Bank. They then focused the METBIND search engine on hCTR1.
They looked for individual atoms in the protein that could be placed within 3.5 Angstrom units of a hypothetical copper ion. One Angstrom unit is equal to one hundred-millionth (10 -8) of a centimeter. They identified six histidine residues in the protein that bind copper (and probably platinum) as the first step in the metal transport process.
They identified nine negatively charged amino acids in the part of the hCTR1 protein that stick out into the extracellular medium waiting for oppositely charged copper or platinum ions to pass by. When the ions arrive, the hCTR1 protein grabs them firmly.
They also found that the hCTR1 trimer creates a neutral channel with a set of triads of methionine amino acids. The triads shepherd copper or platinum ions through the cell membrane into the interior cytoplasm. Each of the methionines is important: if one is lost, copper transport is inhibited. The same effect of methionines has been reported for yeast copper transporter (yCTR).
“Drug developers are interested in the selective transport of platinum and other metal ions into cells to invoke a desired effect, and this study provides a blue print for how they could search for drugs to enhance those effects,” Tsigelny said.
Tsigelny’s research team included Yuriy Sharikov, with SDSC and UC San Diego’s Department of Neurosciences; Jerry P. Greenberg, Mark A. Miller, Valentina L. Kouzentsova, Christopher A. Larson, all with SDSC; and Stephen B. Howell, a professor of medicine at the UC San Diego School of Medicine and associate director for clinical research at the UC San Diego Moores Cancer Center.
The research was funded by grant # W81XWH-08-1-0135 from the National Institutes of Health, as well as grants from the Department of Defense and the Clayton Medical Research Foundation. Computational resources were provided by SDSC and additional support was provided by the UC San Diego Neuroscience Microscopy Shared Facility and the UC San Diego Moores Cancer Center.
About SDSC
As an Organized Research Unit of UC San Diego, SDSC is considered a leader in data-intensive computing and all aspects of ‘big data’, which includes data integration, performance modeling, data mining, software development, workflow automation, and more. SDSC supports hundreds of multidisciplinary programs spanning a wide variety of domains, from earth sciences and biology to astrophysics, bioinformatics, and health IT. With its two newest supercomputer systems, Trestles and Gordon, SDSC is a partner in XSEDE (Extreme Science and Engineering Discovery Environment), the most advanced collection of integrated digital resources and services in the world.
Media Contacts:
Rex Graham, SDSC Communications
858 232-2706 or ragraham@ucsd.edu
Jan Zverina, SDSC Communications
858 534-5111 or jzverina@sdsc.edu
Warren R. Froelich, SDSC Communications
858 822-3622 or froelich@sdsc.edu
Categories
Archive
Related Links
UC San Diego: http://www.ucsd.edu/
UC San Diego Department of Neurosciences: http://neurosciences.ucsd.edu/
UC San Diego Moores Cancer Center: http://cancer.ucsd.edu/
Cell Biochemistry and Biophysics: http://www.springer.com/life+sciences/biochemistry+%26+biophysics/journal/12013
National Institutes of Health: http://www.nih.gov/