News
Loyola Study Uses SDSC’s Expanse to Illustrate Peptide Variations
Published December 18, 2023
Neha Srini and Kimberly Mann Bruch, SDSC Communications
In order for human muscles to properly contract and release, a delicate dance between calcium ion and troponin C (TnC) takes place. If the balance between these two falters, muscles – such as the heart – can fail and result in health issues such as cardiac arrest. Understanding this interaction at the atomic level was the focus of a Loyola University study that used National Science Foundation (NSF) ACCESS allocations on Expanse at the San Diego Supercomputer Center (SDSC) at UC San Diego.
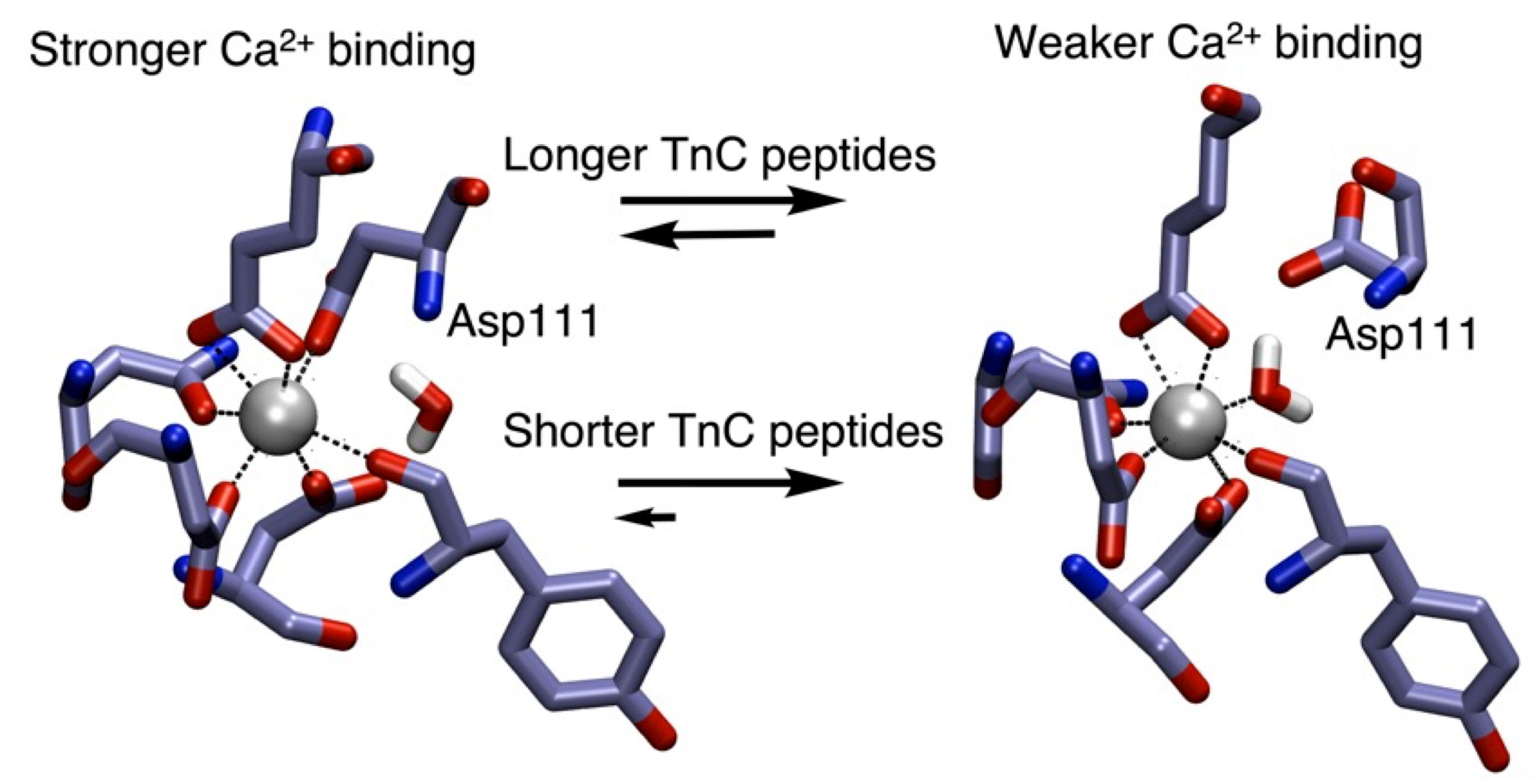
Expanse-generated simulations show how longer troponin peptides have stronger calcium binding than the shorter peptides. Credit: Loyola University Chicago
Led by Loyola’s Department of Chemistry and Biochemistry Assistant Professor Pengfei Li, the team specifically looked at the variations between longer protein peptides and shorter ones. “Our simulations on SDSC’s Expanse indicated that longer peptides maintained a stronger hydrogen-bond network which contributed to more favorable ion binding,” Li explained. “Let’s say that you have a baseball (ion) held by a glove (TnC binding site), but that glove is really old and falling apart so you have a hard time holding onto the ball.”
Li said that the older glove represents shorter peptides, whereby the structural integrity is weak and doesn’t allow for easy binding to the ion. “Longer peptides, on the other hand, can hold onto the ion better because their structural integrity is better maintained by hydrogen bonds,” he said.
Madelyn Smith, who is working on her Ph.D. in chemistry at Loyola, said that using ACCESS allocations on supercomputers like Expanse allowed their team to easily conduct their studies. “Our ultimate goal is to better understand how peptides bind ion and how does the ion-binding influence the subsequent peptide-target binding – working with other research teams that design molecules to regulate these processes,” she said. “Using Expanse enabled us to clearly illustrate the variations between longer peptides and shorter ones.”
The simulation is a prediction of how a troponin peptide behave in water. The overall shape of the troponin peptide is in faded blue, the residues in the troponin/calcium binding site are in bold colors, and the calcium ion is grey. Credit: Madelyn Smith, Loyola University Chicago
The Loyola team’s research was published in Journal of Chemical Information and Modelling in an article titled Molecular Insights into the Calcium Binding in Troponin C through a Molecular Dynamics Study.
The computational research was supported by ACCESS (grant no. BIO210105).

